Computer Vision in Healthcare
Virtual Spatial Biology: Predicting biomarkers for autoimmune disease from standard pathology images.
The Challenge
The "Invisible" Signs of Cancer Risk.
The Eric and Wendy Schmidt Center at the Broad Institute and the Klarman Cell Observatory (KCO) are addressing a critical diagnostic gap in Inflammatory Bowel Disease (IBD). Patients with IBD have chronic inflammation that doubles their risk of colorectal cancer.
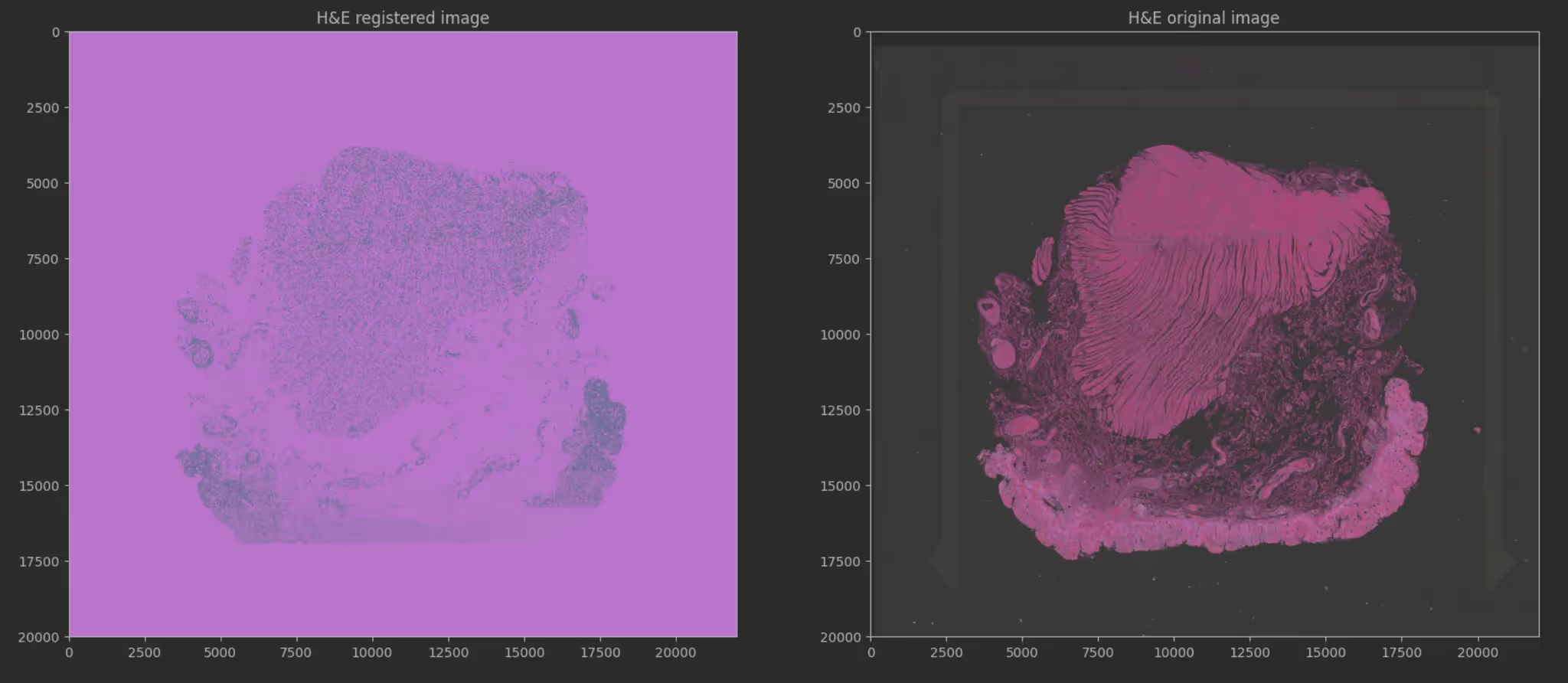
Currently, detecting pre-cancerous lesions (dysplasia) relies on pathologists manually reviewing H&E stained tissue slides. However, molecular changes occur before visual changes. The Broad Institute wanted to use Machine Learning to see these invisible molecular signals. The challenge was finding a way to map standard, affordable tissue images (H&E) to complex, expensive genetic data (Spatial Transcriptomics) to enable mass screening.
Our approach
A Three-Phase R&D Pipeline.
Instead of a simple "accuracy" contest, the Broad Institute and Crunch designed a full R&D pipeline simulation across three phases ("Crunches"):
- Crunch 1 (Prediction): Can models predict the expression of 460 specific genes just by looking at a tissue image?
- Crunch 2 (Scale/Imputation): Can those models generalize to infer a much larger panel of 2,000 genes (unseen data), mirroring clinical scaling?
- Crunch 3 (Discovery): Can participants use these predictions to rank the top 50 gene markers that distinguish pre-cancerous tissue from healthy tissue?
This structure moved beyond "model building" to "biomarker discovery."
The Solution
Multimodal Foundation Models.
The challenge attracted nearly 1,000 experts from 62 countries. The winning solutions (from researchers at ETH Zürich, Stanford, and Mayo Clinic) deployed advanced Multimodal Learning techniques to bridge the gap between vision and biology.
The Methodology:
- Histopathology Foundation Models: Top performers leveraged foundational image encoders (trained on massive pathology datasets) to extract deep visual features from the tissue slides.
- Contrastive Learning: Winner Alexis Gassmann used contrastive learning to align image data and molecular data in a shared latent space, allowing accurate transfer from single-cell data to spatial contexts.
- Spatial Attention: Teams used "Gaussian masking" and "Successive Attention" to model not just the cell itself, but how the tissue architecture around the cell influences gene expression.
Some of the approaches that stood out:
- DeepSpot Enhancement: Kalin Nonchev from ETH Zurich extended the DeepSpot methodology to support 10x Genomics Xenium data, dramatically improving single-cell gene expression predictions.
- Contrastive Learning: Alexis Gassmann applied contrastive learning to build shared embedding spaces between pathology images, gene expression data, and spatial coordinates.
- Creative Proxy Supervision: Team Cellmates solved the fundamental problem of predicting genes with no training data by using FAISS algorithm to find similar single-cell samples and creating supervised learning from unsupervised data.
- Vision-Attention Architecture: The team from IIT Ropar combined vision transformers with multi-head attention networks, demonstrating how modern AI architectures could tackle complex biological problems.

Kalin Nonchev from ETH Zurich's Department of Computer Science secured first place with his DeepSpot methodology.
DeepSpot's application generated 1,792 spatial transcriptomics samples from The Cancer Genome Atlas (TCGA) cohorts, analyzing 37 million spots across melanoma and renal cell carcinoma datasets. The methodology demonstrated multi-cancer validation across metastatic melanoma, kidney, lung, and colon cancers, achieving significant improvement in gene correlation compared to existing methods.
Following the conclusion of the challenge in March 2025, Schmidt Center scientists analyzed the top-performing models and ordered a custom dysplasia gene panel based on AI predictions for cancer detection. Custom gene panels are now in manufacturing, with wet-lab validation experiments launching soon. Winners will be announced in December 2025.

This challenge demonstrates how introducing the global machine learning community to a biological problem can accelerate scientific and clinical discoveries. Looking beyond the boundaries of one domain reveals opportunities we wouldn’t find alone.
The Impact
From Code to Clinical Validation.
The challenge successfully validated the concept of "Virtual Spatial Biology"—using AI to infer expensive biological data from cheap images.
Key Results:
- Experimental Validation: The top 50 gene markers identified by the winning models in Crunch 3 are currently being tested in patient samples via wet-lab experiments at the Broad Institute.
- Clinical Translation: The project demonstrated a pathway to replace expensive spatial transcriptomics with AI-enhanced standard imaging, potentially lowering the cost of cancer screening by orders of magnitude.
- Interdisciplinary breakthrough: The challenge proved that "outsider" ML experts could adapt architectures meant for other domains (like computer vision) to solve highly specific biological problems in IBD.

